This is excellent news! The big funding bill Bush just signed into law includes the provision that all NIH funded research be made open access within 12 months of publication.
Read more here.
Thursday, December 27, 2007
Thursday, August 09, 2007
PCR
 It is an exciting time to be following progress in the field of microbial ecology. While the awareness of the abundance of microscopic organisms is not new, the development of new methods to observe the microscopic world continue to deepen our understanding. Some of these methods have been described briefly on this blog. See the entry on 454 sequencing for example.
It is an exciting time to be following progress in the field of microbial ecology. While the awareness of the abundance of microscopic organisms is not new, the development of new methods to observe the microscopic world continue to deepen our understanding. Some of these methods have been described briefly on this blog. See the entry on 454 sequencing for example.One of the most important techniques for modern microbial ecology and biology as a whole is the polymerase chain reaction or PCR. I suspect most of my modest readership is familiar with the technique, if any of you are not, it is something that you ought to take the time to learn about. A google search of the term PCR reveals a large number of pages devoted to explaining the technique. Many people have also developed diagrams and animations to aid in the understanding. The problem is, most of the good animations do not stand on their own. To make use of them, some background knowledge is needed.
The best animation I have seen on the web is this one. Go ahead and look at both the amplification animation and the interactive graph showing the number of copies of the target molecule present after each cycle.
Here are some things to keep in mind:
- DNA is a double stranded molecule and the two strands are held together by hydrogen bonds. Each individual bond is weak, the strength of the bonding between the two strands arises from the sheer number of individual bonds present. Key point for PCR: The bonding that holds the double strand together is easily disrupted by heat. Thus the 95 deg C steps
- DNA is made up of 4 nucleotides: adenine, thymine, cytosine and guanine attached to each other along a sugar-phosphate backbone. The hydrogen bonding between the two strands of the double stranded molecule are between these nucleotides. The pairing of the nucleotides is specific. Adenine always binds with thymine and guanine with cytosine. Key point for PCR: knowing the sequence of one of the strands of the double stranded DNA makes it possible to deduce the sequence of the opposite strand.
- DNA polymerase is the enzyme required for PCR. The enzyme is capable of synthesizing double stranded DNA from single stranded DNA using the single strand as a template. The activity of this enzyme is specific in several ways. Most importantly for PCR:
- The nucleotide bases strung along the sugar-phosphate backbone of each DNA strand has directionality.a and each of the two strands in the double stranded molecule are oriented in the opposite direction. DNA polymerase can elongate in only one direction. Key point for PCR: the DNA polymerase must elongate each of the two strands from opposite ends.b
- DNA polymerase can not elongate single stranded DNA. A short fragment of double stranded DNA is needed. Key point for PCR: Small lengths of double stranded DNA need to be created flanking the region targeted for amplification (these are the primers).
- The exponential nature of PCR amplification depends on multiple cycles of amplification involving both strands of the double stranded molecule. Key point for PCR: Two primers are needed.
- DNA polymerase - the enzyme.
- 2 primers - Small fragments of single stranded DNA. These are used to produce short regions of double stranded DNA flanking the sequence targeted for amplification.
- Free nucleotides - These must be in the form of nucleotide triphosphates. In this form, they provide the source of the bases needed to build new strands of DNA and the energy required to drive the reaction.
- Template - some DNA containing the region that is to be targeted.
There is much more to say about PCR but this post is already long enough so go enjoy the animation.
aFor a better understanding of the structure of the DNA molecule itself see DNA is a polynucleotide by Larry Moran at Sandwalk.
bSee here and here for an interesting exception to this rule on directionality.
Image from Flickr
Wednesday, August 08, 2007
Bacteria and the cost of oil
 Oil is considered sour if it contains reduced sulfur compounds (sulfides) at concentrations of 1% or greater. High concentrations of sulfides in oil are problematic for a variety of reasons including:
Oil is considered sour if it contains reduced sulfur compounds (sulfides) at concentrations of 1% or greater. High concentrations of sulfides in oil are problematic for a variety of reasons including:- Hydrogen sulfide is extremely corrosive and can cause damage to the pipes used to transport oil.
- When complexed with other metals such as iron, the sulfides can form precipitates that restrict the flow of oil in the pipes.
- Sulfides are toxic and cause environmental and health problems in areas where sour oil is produced, processed or burned.
Souring of oil is exacerbated by the common practice of pumping water into older oil fields to increase the pressure in the fields as a way to increase oil recovery. Depending on the source of the water used, this practice can introduce large quantities of sulfate (SO4) into the oil/water mixture. Any oxygen present in the water when it is first pumped underground is rapidly consumed by microbial activity. Once the oxygen is gone, anaerobic microbes can contiune to extract energy from the organic mater present by using compounds other than oxygen as terminal electron acceptors. Sulfate reducing bacteria or SRBs are anaerobes that are able to use sulfate as an electron acceptor. The process results in the production of oxidized carbon compounds and reduced sulfur (sulfides). Biocides are often added to the water to inhibit microbial activity but this process is not efficient requiring enormous amounts of toxic compounds to be added to the water to have a lasting impact.
Thus, microbial activity in oil fields contributes the the cost of oil production. Microbial sulfide production is not limited to oil reservoirs. SRBs are also responsible for the 'rotten egg' smell associated with other anaerobic environments such as swamps and septic systems.
Thursday, July 19, 2007
Kofi Annan's missed opportunity
 Here is one of the many articles this week in overseas papers covering the announcement that the new organization, Alliance for a Green Revolution in Africa or AGRA led by the former UN chief Kofi Annan will attempt to engineer a green revolution in Africa without the aid of genetically modified (GM) crops. This decision is very short sighted.
Here is one of the many articles this week in overseas papers covering the announcement that the new organization, Alliance for a Green Revolution in Africa or AGRA led by the former UN chief Kofi Annan will attempt to engineer a green revolution in Africa without the aid of genetically modified (GM) crops. This decision is very short sighted.From the article:
Conventional methods of farming have not yet been applied to the fullest extent in Africa. Simply working with conventional breeding, we can do a lot,' said Joseph De Vries, programme director with AGRA.Yes, but, with GM crops, even more could be done. I understand that GM crops are controversial and many people find their use disturbing. However, on a continent where so many go hungry, closing the door completely on a technology that has the potential to improve the drought and pest resistance of important crops makes no sense. One of AGRA's primary goals is to improve "crop varieties for larger, more diverse, and more reliable harvests". How can anyone suggest that in this day and age, GM crops have no role to play in this endeavor?
The genie is out of the bottle. GM crops are here to stay. They should stay. On a planet with 6 billion people and counting, the potential they offer to increase yields, reduce chemical usage and expand arable land is too great to ignore.
The big challenge with the development of GM crops (and the aspect that I am most uncomfortable with) is that too many decisions about which traits to manipulate and what risks are worth taking are made by big agribusiness. This is where Annan's new organization could have played a constructive role. AGRA is headed by a former Secretary-General of the UN and bankrolled by the Gates and Rockefeller foundations to the tune of $150 million. Such an organization has the potential to be a powerful voice in the debate over the best use of GM crops for improving the quality of life and sustainability of agriculture in Africa.
By this decision, AGRA has removed itself a discussion that will occur whether they chose to participate or not.
Wednesday, July 18, 2007
Gecko/mussel hybrid velcro
This looks cool:
A reversible wet/dry adhesive inspired by mussels and geckos
Lee, Lee & Messersmith
Nature 448, 338-341 (19 July 2007)
From the abstract:
A reversible wet/dry adhesive inspired by mussels and geckos
Lee, Lee & Messersmith
Nature 448, 338-341 (19 July 2007)
From the abstract:
Researchers have attempted to capture these properties of gecko adhesive in synthetic mimics with nanoscale surface features reminiscent of setae; however, maintenance of adhesive performance over many cycles has been elusive and gecko adhesion is greatly diminished upon full immersion in water. Here we report a hybrid biologically inspired adhesive consisting of an array of nanofabricated polymer pillars coated with a thin layer of a synthetic polymer that mimics the wet adhesive proteins found in mussel holdfasts. Wet adhesion of the nanostructured polymer pillar arrays increased nearly 15-fold when coated with mussel-mimetic polymer. The system maintains its adhesive performance for over a thousand contact cycles in both dry and wet environments. This hybrid adhesive, which combines the salient design elements of both gecko and mussel adhesives, should be useful for reversible attachment to a variety of surfaces in any environment.Check out the gecko images here: http://www.lclark.edu/~autumn/PNAS/
Stability - Diversity relationships
I mentioned in this post, my concerns about speculations by Xu et al on the role of the human host in the maintenance of a diverse gut microbial community. The proposed benefit to us is that the high diversity encouraged stability and assured that our guts continued to provide the desired services, but the mechanism by which we control diversity was not clear.
An article by Ives and Carpenter
Understanding the dynamics of complex systems such as the human gut is challenging. Here is the background knowledge Ives and Carpenter suggest is necessary for beginning to develop an understanding of the diversity/stability relationship:
And, here is a excerpt from the recommendations they make at the end of the paper:
An article by Ives and Carpenter
Understanding the dynamics of complex systems such as the human gut is challenging. Here is the background knowledge Ives and Carpenter suggest is necessary for beginning to develop an understanding of the diversity/stability relationship:
Before designing an empirical study, it is necessary to know enough about the dynamics of an ecosystem and the environmental perturbations that impinge upon it to select appropriate definitions of stability; there will often be several appropriate definitions. These concepts also identify key features—we will refer to them as mechanisms—that together dictate stability. These mechanisms involve the strength of interactions among species, the mode in which species interact (whether they are competitors, predators, mutualists, etc.) that gives the food-web topology, and the ways in which species experience different types of environmental perturbations. Because both species interactions and environmental perturbations can drive fluctuations in species densities, these must be sorted out and quantified to understand their mechanistic roles in diversity-stability relationships.
And, here is a excerpt from the recommendations they make at the end of the paper:
The human gut community does exhibit characteristics of a stable system such as the ability to resist perturbations. So, what are the mechanisms that maintain the diversity, what is the diversity stability relationship and how do we go about studying it.The relationship between diversity and stability has interested ecologists since the inception of the discipline (35), and the absence of a resolution reflects the complexity of the problem. Much of the complexity derives from the multiplicity of diversity-stability relationships, depending on the definitions of diversity and stability and on the context in which an ecosystem is perturbed. We cannot expect a general conclusion about the diversity-stability relationship, and simply increasing the number of studies on different ecosystems will not generate one.
Rather than search for generalities in patterns of diversity-stability relationships, we recommend investigating mechanisms. A given diversity-stability relationship may be driven by multiple mechanisms, and the same mechanisms may evoke different diversity-stability relationships depending on the definitions of diversity and stability. We need more studies revealing exactly what these mechanisms are. This requires models joined to empirical studies that can reproduce, in a statistically robust way, not only a diversity-stability relationship but also the dynamics exhibited by a system.
Monday, July 09, 2007
More on human guts
Yet another interesting open access gut microbe paper in PLoS Biology came out in June. This one describes patterns in the colonization of the intenstines of human infants. As mentioned in a previous post, we are born with a sterile intestinal tract and depend upon the ingestion of compatible microbes for the establishment of our gut community. This study used 16S rhibosomal DNA sequences to document changes in the structure and diversity of infant guts over the first year of life. As with the previous paper, Liza Gross wrote a nice summary article.
Some key points:
This paragraph from the end of the Gross summary provides a good overview of the most interesting findings:
Some key points:
- 14 babies were followed (including one set of twins) for one year. Early the communities were quite different but by the end of the first year they had acquired a composition similar to that of the adult human.
- At one week of age, two babies delivered by cesarian had fewer total gut bacteria indicating that during natural child birth, the colonization begins during the birthing process.
- While broadly similar to each other and to the adult community, each infant had a distinct profile that persisted over time.
This paragraph from the end of the Gross summary provides a good overview of the most interesting findings:
The idiosyncratic nature of the early stages of colonization suggests that a baby’s initial bacterial profile largely results from incidental microbial encounters. The fact that some of the early stool samples matched their mother’s breast milk or vaginal sample supports this interpretation. Shared environment may also explain the coincidental appearance of microbes in the twins. The researchers explain the tendency of these communities to eventually converge by hypothesizing that the human–microbe symbiosis has likely evolved under strong selection and that certain well-adapted microbes repeatedly “win” the battle over the opportunistic early colonizers.Selections from the final paragraph describes some of the future directions the work will take:
By comparing the surprising range of microbial profiles found in these healthy babies to the microbiota of infants born prematurely or with health problems, future studies can explore how diet, delivery method, or other factors might spell the difference between health and disease.and that the approach used in the study will allow us to explore questions about
the environmental and genetic factors that shape and personalize the amazing “alien” ecosystem that lives within us.
Saturday, July 07, 2007
Bacteriorhodopsin phototrophy
 Classification of the metabolic capabilities of microbes can be challenging. With few exceptions, macroorganisms are either photosynthesizing primary producers (photo-autotrophs) or consumers (organotrophs or more commonly, heterotrophs).
Classification of the metabolic capabilities of microbes can be challenging. With few exceptions, macroorganisms are either photosynthesizing primary producers (photo-autotrophs) or consumers (organotrophs or more commonly, heterotrophs).For microbes, the story is more complicated. In addition to phototrophy, microbes can be chemo- or litho-trophs meaning they are able to derive energy from the oxidation of inorganic compounds such as reduced sulfur. If they can use light or chemical energy to fix carbon, then they are considered autotrophs. If the energy they acquire can be used to synthesize ATP but not to fix carbon, they are dependent on external sources of organic carbon making them mixotrophs.
An example of a phototrophic mixotroph is pictured above. These are salt loving haloarchaea in salt production ponds near (in?) San Francisco. The red color is due to the transmembrane protein bacteriorhodopsin. Using this protein some haloarchaea can harness sunlight to pump protons across their cell membrane. This establishes a proton gradient across the membrane. This gradient can be used to generate ATP.
There is a large number of scientific papers on bacteriorhodopsin because of its relative simplicity, it has become a model system for the study of membrane associated ion pumps.
Image from here http://www.sfgate.com/cgi-bin/article.cgi?file=/chronicle/archive/2001/03/13/MN152047.DTL
These salt ponds are near San Francisco. If you want another view of the bay area ponds follow this link, select the satellite map and zoom way in. I tried this for a few of the other places I know these salt production ponds exist but the satellite images did not provide good enough resolution. An example is Bon Aire in the Netherlands Antillies
Labels:
microbiology,
mixotrophy,
Molecular Biology,
Science
Friday, July 06, 2007
Friday dog post?
This one is for ERV
 This photo won us a years supply of dog food at a local fundraiser a few years ago. The muzzle has gotten whiter since then. It may be due to the fact that he now shares the house with two young kids.
This photo won us a years supply of dog food at a local fundraiser a few years ago. The muzzle has gotten whiter since then. It may be due to the fact that he now shares the house with two young kids.
 This photo won us a years supply of dog food at a local fundraiser a few years ago. The muzzle has gotten whiter since then. It may be due to the fact that he now shares the house with two young kids.
This photo won us a years supply of dog food at a local fundraiser a few years ago. The muzzle has gotten whiter since then. It may be due to the fact that he now shares the house with two young kids.
Thursday, July 05, 2007
In The Bay 3 July 2007 II
Sustainable education
The most recent issue of the journal Nature has a review (behind a subscription barrier) of a new book: Degrees that Matter: Climate Change and the University by Ann Rappaport & Sarah Hammond Creighton. I have not read the book but some of the information is interesting.
The book documents a 15 year effort by Tufts University that began in 1991 to reduce its energy consumption. From the review:
The book documents a 15 year effort by Tufts University that began in 1991 to reduce its energy consumption. From the review:
The central observation from Degrees that Matter is that universities are in a unique position to offer leadership on climate change and carbon emissions through their educational, research and wider roles in society.The conclusion is a bit disheartening:
The bad news is that despite the intense programme, carbon emissions at Tufts — both net and normalized — seem to have increased over time. The university as a whole has become more energy intensive, with the consequence that it will not meet its Kyoto target. This should, however, be set against other higher-education institutions, where the rate of increase over similar time periods is much greater and the reversal of trends, if at all, much slower. A large part of the increase is due to growing demands from personal equipment.I assume the "personal equipment" are computers which consume an enormous amount of energy.
Wednesday, July 04, 2007
In the Bay 3 July 2007
Since I wrote about marine cilliates in my previous post, I thought I'd try to find some for this edition of In The Bay.
This is a group of Vorticella or Vorticella-like stalked cillates. As you watch the movie you can see the feeding current they are creating with their cillia. About 21 seconds into the movie (6 seconds from the end) the whole colony retracts. This is a defensive mechanism and occurs extremely fast.
This is a group of Vorticella or Vorticella-like stalked cillates. As you watch the movie you can see the feeding current they are creating with their cillia. About 21 seconds into the movie (6 seconds from the end) the whole colony retracts. This is a defensive mechanism and occurs extremely fast.
Saturday, June 30, 2007
Cilliate kleptoplasty
 Chloroplasts, like mitochondria, have their own chromosomal DNA. This is, of course, evidence for the endosymbiotic theory of the origin of chloroplasts. It is also useful because it allows researchers to use the DNA to identify the source of the chloroplasts present in kleptoplastic organisms. This is a fairly standard method and is the way that Gast et al 2007 determined the source of the chloroplasts in the Antartic dinoflagellates discussed in a previous post.
Chloroplasts, like mitochondria, have their own chromosomal DNA. This is, of course, evidence for the endosymbiotic theory of the origin of chloroplasts. It is also useful because it allows researchers to use the DNA to identify the source of the chloroplasts present in kleptoplastic organisms. This is a fairly standard method and is the way that Gast et al 2007 determined the source of the chloroplasts in the Antartic dinoflagellates discussed in a previous post.In a paper (pdf) a few years ago in Limnology and Oceanography, McManus et al. used the chloroplasts present in the kleptoplastic tide-pool ciliates, Strombidium oculatum and Strombidium styliferseen to help reveal an interesting life history. The chloroplasts were from the large multicellular macroalgae Enteromorpha clathrata which raised the question of how these unicellular cilliates were able to acquire macroalgal chloroplasts.
McManus et al. found that the cilliates don't appear to be grazing directly on the large strands of the mature algae but on the small motile reproductive cells, called zoospores, mature algal strands release.
In addition to chloroplasts, the zoospores contain a pigmented eyespot. As the photo above (from the paper's figure 1) demonstrates, the kleptoplastic cilliates contain a pigmented eyespot similar to the ones possessed by the zoospores. This suggests that the Strombidium cilliates also owe their phototaxic abilities to the alga cells they ingest.
Some other interesting points about these cilliates:
- They are tidal organisms and live by the rhythm of the tides, becoming active during low tide when tidal pools are calm, and then attach to surfaces and encyst during high tide, presumably to prevent them from being washed out to sea and away from their food.
- They appear to be obligate mixotrophs, unable to grow in the dark or in the absence of algal food.
McManus, G. B., H. Zhang, and S. Lin. 2004. Marine planktonic ciliates that prey on macroalgae and enslave their chloroplasts. Limnol. Oceanogr. 49:308-313.
Tuesday, June 26, 2007
8 random facts meme
I have been tagged by John Dennehy (aka the Evilutionary biologist) in this 8 random facts meme. The rules are:
1. When I saw I'd been tagged for this, my first though was: how big a population of facts do I need to generate in order to make sure that the list of 8 will seem random?
2. I own more that one microscope.
3. I have never known what I wanted to be when I grow up (I still don't).
4. I love the change of seasons and do my best to never complain about the weather.
5. There is a definite trend in the size of the organisms I've studies since college: penguins > tilapia > phytoplankon > bacteria ... ?
6. My dog's name is Larry
7. I live on an island off the the coast (if Spalding Grey can say that about Manhattan, I can say it about Aquidneck Island)
8. I am pretty sure this is not a random list but we'll have to check with Larry to be certain.
So many people have already done this so for the last part, I'll steal this from Tara: anyone who's not been hit already, feel free to consider yourself tagged.
- We have to post these rules before we give you the facts.
- Players start with eight random facts/habits about themselves.
- People who are tagged need to write their own blog about their eight things and post these rules.
- At the end of your blog, you need to choose eight people to get tagged and list their names.
- Don’t forget to leave them a comment telling them they’re tagged, and to read your blog.
1. When I saw I'd been tagged for this, my first though was: how big a population of facts do I need to generate in order to make sure that the list of 8 will seem random?
2. I own more that one microscope.
3. I have never known what I wanted to be when I grow up (I still don't).
4. I love the change of seasons and do my best to never complain about the weather.
5. There is a definite trend in the size of the organisms I've studies since college: penguins > tilapia > phytoplankon > bacteria ... ?
6. My dog's name is Larry
7. I live on an island off the the coast (if Spalding Grey can say that about Manhattan, I can say it about Aquidneck Island)
8. I am pretty sure this is not a random list but we'll have to check with Larry to be certain.
So many people have already done this so for the last part, I'll steal this from Tara: anyone who's not been hit already, feel free to consider yourself tagged.
Monday, June 25, 2007
In the Bay 24 Jun 2007
Thursday, June 21, 2007
Building a better biofuel
 Biofuels offer the promise of reducing our dependence on fossil fuels. The most widely used biofuel is ethanol made by the biological fermentation of corn. This process is not as green as many would like to believe because a substantial amount of energy (in the form of fossil fuel) is used in the production process. So ,the net gain is not great. Also, ethanol itself is not a terribly good fuel as it is very volatile, is not very energy dense and absorbs water.
Biofuels offer the promise of reducing our dependence on fossil fuels. The most widely used biofuel is ethanol made by the biological fermentation of corn. This process is not as green as many would like to believe because a substantial amount of energy (in the form of fossil fuel) is used in the production process. So ,the net gain is not great. Also, ethanol itself is not a terribly good fuel as it is very volatile, is not very energy dense and absorbs water.A better biofuel would be one that does not rely on an important food crop, has a higher energy density and can be produced with as little energy input as possible. In todays issue of Nature, Román-Leshkov et al present a letter in which they report on a process by which the are able to produce 2,5-dimethylfuran (DMF) from the sugar fructose.
DMF is a better fuel than ethanol and interest in it is not new. What is new in this report is the ability to produce DMF in an industrial process requiring much less energy than previously reported methods.
The environmental impacts of this material have not been well studied and the source of fructose for the production of DMF remains an important issue but this type of innovative thinking has a place in our efforts to move away from dependence on fossil fuels.
Complete citation:
Roman-Leshkov, Y., C. J. Barrett, Z. Y. Liu, and J. A. Dumesic. 2007. Production of dimethylfuran for liquid fuels from biomass-derived carbohydrates. Nature 447:982-985.
Cow burps and global warming
My friend (and a reader of this blog) Marek Kirs send me this link to a story on NPR about attempts to reduce the amount of methane produced by cows. The idea is to adjust the types of food they eat or even manipulate their gut microbial community to eliminate the methanogenic organisms responsible for the gas production.
One of the points the researchers make is that the production of methane is the result of incomplete oxidation of food eaten by the cows. So, the elimination of methanogenesis could result in increases production of meat and milk from these animals. Cattle food conversion ration are around 12%, and any increases would likely be small but the idea is to have enough of an increase to offset any increased cost to the producers.
Wednesday, June 20, 2007
Gut bacteria metabolic diversity
Another paper exploring microbial diversity of the human gut has been released in PLoS Biology. It is open source so go read it for yourself! There is also a summary article on the data intended for a lay audience. The data presented in the paper is very interesting. They sequenced the genomes of two gut bacteria, Bacteroides vulgatus and Bacteroides distasonis, and contrasted their metabolic potential (the genes present in the genome) with the well studied species Bacteroides thetaiotaomicron. This comparison does provide some evidence that niche specialization plays a role in maintaining the diversity of the gut microbiota.
The importance of studying the gut microbiota is summed up nicely in the beginning of the author summary:
I would have worded the first sentence differently as saying 'have not had to" sounds a bit too teleological to me.
I am also troubled by their use (later in the introduction) of the term "top-down selection" in reference to host driven selective forces that they argue are responsible for maintaining a high degree of functional redundancy in the gut community. I am not aware that the term top-down selection, as used by the broader ecology community, is considered a force for the maintenance of ecosystem stability.
The end of the paper lists a set open questions that it would be very nice to have answers to:
The importance of studying the gut microbiota is summed up nicely in the beginning of the author summary:
"Our microbial partners provide us with certain features that we have not had to evolve on our own. In this sense, we should consider ourselves to be a supraorganism whose genetic landscape includes both our own genome as well as the genomes of our resident microbes, and whose physiologic features are a synthesis of human and microbial metabolic traits."
I would have worded the first sentence differently as saying 'have not had to" sounds a bit too teleological to me.
I am also troubled by their use (later in the introduction) of the term "top-down selection" in reference to host driven selective forces that they argue are responsible for maintaining a high degree of functional redundancy in the gut community. I am not aware that the term top-down selection, as used by the broader ecology community, is considered a force for the maintenance of ecosystem stability.
The end of the paper lists a set open questions that it would be very nice to have answers to:
"Do we share an identifiable core “microbiome”? If there is such a core, how does the shell of diversity that surrounds the core influence our individual physiologic properties? How is the human microbiome evolving (within and between individuals) over varying time scales as a function of our changing diets, lifestyle, and biosphere? Finally, how should we define members of the microbiome when microbes possess pan-genomes (all genes present in any of the strains of a species) with varying degrees of “openness” to acquisition of genes from other microbes?"
Sunday, June 17, 2007
Bacterial predators
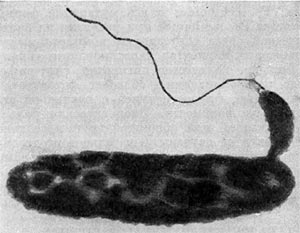 With the rise of antibiotic resistance in important bacterial pathogens, attention is being focused on alternative treatments for bacterial infections. The most prominent among these is phage therapy. Another potential antimicrobial agent is the predatory bacteria Bdellovibrio and related species
With the rise of antibiotic resistance in important bacterial pathogens, attention is being focused on alternative treatments for bacterial infections. The most prominent among these is phage therapy. Another potential antimicrobial agent is the predatory bacteria Bdellovibrio and related speciesI am not sure how likely it is that these bacteria will ever find use as therapeutic agents, but they are pretty amazing organisms and they definitely have the potential to be used in this way.
Bdellovibrio have a truly predatory life style. They require specific species of gram negative bacteria to grow. The cells are small and highly motile. When they encounter a susceptible host cell, they attach themselves to the outer surface of the cell, bore a hole in the cell wall and push themselves into the periplasmic space (the space between the outer membrane and cytoplasmic membrane of gram negative bacteria). Once there the cell is killed and its contents consumed. Bdellovibrio reproduces within the cell and once the contents of the prey are consumed, the daughter cells will burst out of the shell of the dead prey cell and head off to find other targets.
Thursday, June 14, 2007
454

DNA sequencing has revolutionized biology. Not too long ago obtaining the sequence of an individual gene was a big deal. Now sequencing entire genomes is becoming routine. To obtain the sequence of an organism's entire genome, the genome is broken into fragments and many many fragments are sequenced individually using a PCR based method. In the standard process, the sequence generated from each fragment is about 500 to 1,000 bases long. To generate the sequence of the genome, computers are used to align all of the data and identify contiguous sequences that can then be assembled. In order to be confident in the data (errors are made in the process), the entire genome is sequenced multiple times. At least 4 to 5 x coverage is considered necessary for good data.
A typical bacterial genome of 3.5 mega-bases sequenced to 5x coverage amounts to over 21,000 individual sequencing runs. For organisms (such as humans) which have multiple chromosomes each chromosome is sequenced individually. With about 3 billion bases in the human genome and 10x coverage, the data contained in the human genome project represents an enormous amount of sequencing and alignment effort.
A relatively new innovation called 454 sequencing has sped up the process. 454 is still based on PCR but it is much faster as the sequence is read as the PCR progresses. The trade off to the increased speed is that each individual read is somewhat shorter making assembly more challenging.
One 454 application that is becoming more and more widespread is the sequencing of multiple strains of a species of bacteria. In this type of application the genome of one strain is already known. The assembly of the genome of each successive strain is facilitated by the use of the original strains sequence as a template.
454 sequencing was used to determine James Watson's genome. The whole genome was assembled in two months at a cost of $1 million. The assembly was almost certainly accelerated by aligning short reads of Watson's DNA with the pre-existing human genome sequence.
image from the tutorial found on 454 life sciences web page
Monday, June 11, 2007
Kleptoplasty

Kleptoplasty is a wonderful term. It is used to describe the behavior of a group of organisms that are able to ingest algal cells and degrade the cells, but not the chloroplasts contained within the cells. The chloroplasts remain functional for some period of time during which the photosynthetic products generated by the sequestered chloroplasts are utilized by the new 'host'.
On the left is a figure from a paper by Gast et al. in the journal Enviornmental Microbiology from earlier this year showing a kleptoplastic dinoflagellate isolated from the Ross Sea in Antartica (the paper is in a free issue of the journal so go read the whole thing).
In addition to being a very interesting behavior from an ecological perspective, kleptoplasty is of evolutionary interest because the capacity to grow autotrophically by photosynthesis arose within dinoflagellates by the retention of chloroplasts from ingested algal cells. This ability appears to have arisen multiple times within dinoflagellates because not all contain chloroplasts from the same type of algal cell.
Sunday, June 10, 2007
African deforestation
 The rainforest of the Congo Basin in Central Africa is the second largest in the world. It covers millions of square kilometers and spans 6 different countries (including Gabon where I spent 2 years). As with other tropical rain forests this region is home to substantial biodiversity. Here in the US we hear a great deal about deforestation in the Amazon, less is heard about the African rainforests.
The rainforest of the Congo Basin in Central Africa is the second largest in the world. It covers millions of square kilometers and spans 6 different countries (including Gabon where I spent 2 years). As with other tropical rain forests this region is home to substantial biodiversity. Here in the US we hear a great deal about deforestation in the Amazon, less is heard about the African rainforests.The most recent issue of the journal Science contains a brief report documenting the extent of deforestation and increases in commercial logging in the Congo Basin. The information was acquired by analyzing almost 30 years of satellite data for changes in forest cover and the appearance of new logging roads. The objectives of the report were to highlight the lack of good data on the extent of deforestation in the Congo Basin, present the data acquired from the available satellite data and demonstrate the value of the satellite data as a tool to assess changes in land use and document the extent of logging.
*added on edit: Here is some more information about the article and here is the figure from the paper showing the extent of the deforestation. Note how much of the area still has >75% forest cover.
Saturday, June 09, 2007
adopt a cheetah
A recent report in Proceeding of the Royal Society describes the genetic relationship among cheetah litter mates. They found that almost half the litters were comprised kittens of mixed paternity. Female cheetah parole a territory of 300-400 km2 and this allows them to interact with more than one male during a single reproductive cycle. This is a behavior that they share with the common cat. A summary of the paper can be found here.
One finding that struck me was that within the 47 litters surveyed, they found 3 instances of adoption by mother cheetahs of unrelated kittens. This finding has been described previously and there are several ideas as to the potential selective advantage this behavior. My first question is given the huge territories these cats parole, how on earth do the new mothers find orphaned kittens?
One finding that struck me was that within the 47 litters surveyed, they found 3 instances of adoption by mother cheetahs of unrelated kittens. This finding has been described previously and there are several ideas as to the potential selective advantage this behavior. My first question is given the huge territories these cats parole, how on earth do the new mothers find orphaned kittens?
Thursday, June 07, 2007
Plant-bacteria communication
In a previous post I mentioned that the nitrogen fixation performed by leguminous plants is the result of a symbiosis between the plants and bacteria. In this relationship, the plants create an environment within their roots (called nodules) where the bacteria take up residence. In exchange for energy provided by the plant, the bacteria fix atmospheric nitrogen that is then made available to the plant. Allowing bacteria access to the interior of the root is a potentially risky act on the part of the plant. So, a mechanism to exclude all bacteria except for the desired nitrogen fixer has developed and the symbiotic relationships are species specific. Each plant has specific bacterial symbionts. In the early stages of the establishment of the root nodule, signals are exchanged between the plant roots and bacteria in the soil. First, the plants release phytochemicals that attract the desired bacteria. The target bacteria are attracted by the phytochemicals and these chemicals induce the bacteria to release their own compounds called nod factors. The nod factors, in turn are sensed by the plant. Detection of the appropriate nod factor will induce the plant to initiate the process of allowing the bacteria access to the interior of the root where the nodule will form.
In the latest PNAS, Fox et al. report that many common pesticides inhibit the formation of root nodules by interfering with communications between the plants and bacteria.
While the significance of this in the environment is not known, it has the potential to be of concern because the use of crop rotations that include plants (such as soy or alfalfa) that are capable of enriching the nitrogen content of the soil is one important way to reduce the use of fertilizer (and energy) in modern agriculture.
In the latest PNAS, Fox et al. report that many common pesticides inhibit the formation of root nodules by interfering with communications between the plants and bacteria.
While the significance of this in the environment is not known, it has the potential to be of concern because the use of crop rotations that include plants (such as soy or alfalfa) that are capable of enriching the nitrogen content of the soil is one important way to reduce the use of fertilizer (and energy) in modern agriculture.
Wednesday, June 06, 2007
In the bay 6 June 2007
One word: SNAILS! They were abundant last time and there are even more of them out there this week.
I know a fair amount about phytoplankton and am pretty good at identifying them. See the list of previous students here. When I came up with the idea of posting microscope images take of samples from the bay here on mixotrophy, I was ( and still am) planning on focusing on phytopankton. Of course this time of year, the zooplankton population is high and all of these little heterotrophs are busy grazing on the phytoplankton. This keeps phytoplankton cell abundance low. As a result, I am not seeing much of what I was planning on photographing. There are plenty of zooplankton to look at of course. The problem is, identifying zooplankton is much harder for me. Getting good live pictures is also a challenge because they move around.
I did get a few nice images though. Here is another larvae viewed in bright field. It is the nauplius stage of some type of crustation (I think).
This is the same view in dark field:

I know a fair amount about phytoplankton and am pretty good at identifying them. See the list of previous students here. When I came up with the idea of posting microscope images take of samples from the bay here on mixotrophy, I was ( and still am) planning on focusing on phytopankton. Of course this time of year, the zooplankton population is high and all of these little heterotrophs are busy grazing on the phytoplankton. This keeps phytoplankton cell abundance low. As a result, I am not seeing much of what I was planning on photographing. There are plenty of zooplankton to look at of course. The problem is, identifying zooplankton is much harder for me. Getting good live pictures is also a challenge because they move around.
I did get a few nice images though. Here is another larvae viewed in bright field. It is the nauplius stage of some type of crustation (I think).

This is the same view in dark field:

Tuesday, June 05, 2007
Computing power!
Check out this video on meta-imaging from a recent TED conference. Phil Plait at BA was struck by the reconstruction of Notre Dame and its potential for use within astronomy. The whole presentation is incredible. I wonder how powerful a computer he was using. With image resolution so high the memory needs must also be huge. Here is a link to the photosynth software. Of course I am using a mac so I can't download it.
Monday, June 04, 2007
Nitrogen fixation
Industrial agriculture is highly dependent on a ready supply of labile nitrogen fertilizer for high yields. Roughly half of all the nitrogen used in agriculture is in the form of ammonia (NH3) synthesized using the Haber-Bosch process. In this process, atmospheric nitrogen in the form of N2 gas is mixed with hydrogen gas at high temperature (500 oC) and pressure (200 atm) in the presence of an iron catalyst. That's 500 oC and 200 atm. Very hot and very high pressure.
Bacteria (and archaea) are also capable of converting nitrogen gas to ammonium (often referred to as 'fixing nitrogen') but they do it at normal temperatures and under 1 atmosphere of pressure. In these organisms the process is also energy intensive but the energy is supplied in the form of ATP and the catalyst is an enzyme complex containing 2 proteins, dinitrogenase and dinitrigenase reductase. This capacity is found in both bacteria and archaea in diverse environments including the root nodules of so called nitrogen fixing plants such as clover and soy. Bacteria, not the plants, fix the nitrogen.
There are many blog worthy aspects of microbial nitrogen fixation. I have created a label for this topic and intend to explore some of them here in the future.
Bacteria (and archaea) are also capable of converting nitrogen gas to ammonium (often referred to as 'fixing nitrogen') but they do it at normal temperatures and under 1 atmosphere of pressure. In these organisms the process is also energy intensive but the energy is supplied in the form of ATP and the catalyst is an enzyme complex containing 2 proteins, dinitrogenase and dinitrigenase reductase. This capacity is found in both bacteria and archaea in diverse environments including the root nodules of so called nitrogen fixing plants such as clover and soy. Bacteria, not the plants, fix the nitrogen.
There are many blog worthy aspects of microbial nitrogen fixation. I have created a label for this topic and intend to explore some of them here in the future.
Sunday, June 03, 2007
Ancient life?
 An item in a list of news reports over at Primordial Blog caught my eye this evening. It is a report in CBC News about a paper in the most recent issue of the journal Geology describing what is believed to be ichnofossils in 3.5 billion years pillow basalts from Australia.
An item in a list of news reports over at Primordial Blog caught my eye this evening. It is a report in CBC News about a paper in the most recent issue of the journal Geology describing what is believed to be ichnofossils in 3.5 billion years pillow basalts from Australia.Ichnofossils, or trace fossils are not the remains of organisms themselves but tracks left by organisms that become preserved. In this case the traces are tunnels in the volcanic rock presumably left by microorganisms as they degraded the rock to mobilize nutrients contained within. The images on the left are from a 1998 report (Fisk, M. R., Giovannoni, S. J. & Thorseth, I. H. (1998) Alteration of Oceanic Volcanic Glass: Textural Evidence of Microbial Activity. Science, 281, 978-980.) describing these tunnels from oceanic basalts. The scale bars are 50 and 10 microns long in the top and bottom panels respectively.
There is some controversy over whether these tunnels are due to the activity of microbes or are the result of some poorly understood form of abiotic chemical weathering. These images are not easy to get as the rock has to be cut into think slices with a diamond saw and then polished to allow light through. The sections that were used to take the images shown here were 30 microns thick.
Saturday, June 02, 2007
MDR mobility
There is much talk in the blogsphere about this idiot who knowingly exposed a huge number of people to a multi drug resistant strain of TB. f you are interested, Tara Smith over at Aetiology has several posts (1,2,3 & 4 so far) on the topic including links to other discussions and information.
Instead of commenting on that situation I thought I'd call attention to a recent paper by Tim Welch and others in Plos One about a plasmid inferring multi drug resistance that has been found to be presentquite common in our food supply.

This plasmid is of concern because it was isolated from a strain of the plague bacteria, Yersinia pestis, back in the mid 1990s. Since then it has not turned up in Y. pestis again but this new paper reports finding related plasmids in the fish pathogen Yersinia ruckeri and in a strain of the food born pathogen Salmonella. The picture on the left shows the relationship between the three plasmids.
The inner circle shows the conserved backbone indicating that the plasmids isolated from three different species of bacteria have a recent, common origin. They all have the genes necessary to be self mobilizing but they differ in the number of drug resistance genes they carry, ranging from 9 to 13.
using data from the conserved backbone Welch et al. screened a large number of MDR strains of Salmonella, E. coli and Klebsiella sp. isolated from agricultural products and found that the majority of the resistant strains harbor a relative of this plasmid.
From the paper:
Instead of commenting on that situation I thought I'd call attention to a recent paper by Tim Welch and others in Plos One about a plasmid inferring multi drug resistance that has been found to be present
This plasmid is of concern because it was isolated from a strain of the plague bacteria, Yersinia pestis, back in the mid 1990s. Since then it has not turned up in Y. pestis again but this new paper reports finding related plasmids in the fish pathogen Yersinia ruckeri and in a strain of the food born pathogen Salmonella. The picture on the left shows the relationship between the three plasmids.
The inner circle shows the conserved backbone indicating that the plasmids isolated from three different species of bacteria have a recent, common origin. They all have the genes necessary to be self mobilizing but they differ in the number of drug resistance genes they carry, ranging from 9 to 13.
using data from the conserved backbone Welch et al. screened a large number of MDR strains of Salmonella, E. coli and Klebsiella sp. isolated from agricultural products and found that the majority of the resistant strains harbor a relative of this plasmid.
From the paper:
The discovery of these MDR IncA/C plasmids in evolutionarily distinct pathogens attests to recent genetic exchange, either directly between these bacterial species or through bacterial intermediates, and it suggests that overlap in the ecological niches of these organisms is sufficient to permit past or future plasmid transmission.And
we present evidence that a common plasmid backbone is prevalent among E. coli, Klebsiella sp. and multiple Salmonella serotypes isolated from retail meats in the US, and among some food animal isolates of E. coli. Our data imply that high levels of MDR in the causative agent of plague may rapidly evolve naturally...
Friday, June 01, 2007
In the bay 30 May 2007 II
Snail larvae. The water was loaded with these. Probably the periwinkle Littorina littorea. An invasive species introduced to Canada from Europe in the 1800. It is abundant on rocky shores throughout New England. I am not sure how far south they are found.
Here are 2 pictures:

Side view of the shell. the blurring at the top right of the image is due to the movement of cilia along the edge of their feeding apparatus called a velum.
End on view:

the picture is fuzzy but you can see the extended velum and the cilia along the edge.
And a video:
Here are 2 pictures:

Side view of the shell. the blurring at the top right of the image is due to the movement of cilia along the edge of their feeding apparatus called a velum.
End on view:

the picture is fuzzy but you can see the extended velum and the cilia along the edge.
And a video:
Thursday, May 31, 2007
Pictures
I enjoyed spending time taking pictures on the microscope last night and hopefully some readers will also enjoy seeing what is in the bay from time to time. I don't have DIC on my little scope so don't expect to be seeing any thing like this.
I actually saw a relative of these last night but they were attached to an ostracod and it was moving too fast for me to get a picture. Check out all of the great pictures at the Olympus site, and anyone interested in contributing to my new microscope fund, please drop me a line.
I actually saw a relative of these last night but they were attached to an ostracod and it was moving too fast for me to get a picture. Check out all of the great pictures at the Olympus site, and anyone interested in contributing to my new microscope fund, please drop me a line.
Wednesday, May 30, 2007
In the bay 30 May 2007*

The marine diatom
Asterionellopsis glacialis
Check out some better picures here and here.
*I am still experimenting with sample collection locations and and I hope to get access to a better microscope/camera setup, but I intend to make "In the bay" a regular feature of the blog. This image was taken on an old Bausch & Lomb compound microscope at ~100x with an inexpensive 0.3 megapixel digital camera. Water was collected with a fine meshed plankton net in a shallow cove on the north end of Aquidneck Island in Narragansett Bay, RI (see the red arrow below).

Tuesday, May 29, 2007
All of us
John Dennehy, the Evilutionary biologist, put up a post last week about an article in Science News reviewing research on the abundance and diversity of microbes associated with the human body. It presents a nice summary. Some fact culled from that article and elsewhere that are worth contemplating:
> There are 10 time (TEN TIMES) as many bacterial cells as mammalian cells in your body. If you like to contemplate zeros the number is estimated to be ~ 1014 bacterial cells to a measly 1013 mammalian.
> The bulk of these microbes live in our gut where densities can reach 1011 to 1012 cells ml-1 making the gut the ecosystem with the highest density of bacterial cells yet described.
> These are diverse communities 400 or so species but with two only 2 divisions (Fermicutes and bacteroides) accounting for 98% of total population)
>As with other microbial communities a very small fraction of the total number of species present are conducive to culture so most of what we know about them is from extracting and sequencing their genetic material.
> At birth the gut is sterile and must be colonized by bacteria ingested from the environment.
I do have one nit to pick with the Science News article. In the section describing the metagenomic efforts directed at sequencing the 'community genome' this is said:
Generating sequence data from the DNA present in a microbial community is useful and worth doing. It provides information about what genes are present and about the metabolic potential of the community. But in isolation, it provides no information about what metabolic activities are actually occurring. The only real way to know what activities are occurring is to measure the activity directly There are practical limitations to doing direct measurements of all enzymatic activities of potential interest so the genetic data is very valuable but its limitations are worth keeping in mind.
> There are 10 time (TEN TIMES) as many bacterial cells as mammalian cells in your body. If you like to contemplate zeros the number is estimated to be ~ 1014 bacterial cells to a measly 1013 mammalian.
> The bulk of these microbes live in our gut where densities can reach 1011 to 1012 cells ml-1 making the gut the ecosystem with the highest density of bacterial cells yet described.
> These are diverse communities 400 or so species but with two only 2 divisions (Fermicutes and bacteroides) accounting for 98% of total population)
>As with other microbial communities a very small fraction of the total number of species present are conducive to culture so most of what we know about them is from extracting and sequencing their genetic material.
> At birth the gut is sterile and must be colonized by bacteria ingested from the environment.
I do have one nit to pick with the Science News article. In the section describing the metagenomic efforts directed at sequencing the 'community genome' this is said:
"Called metagenomics, this form of analysis doesn't produce a list of bacteria but instead describes the metabolic activities going on within a microbial community. These activities include energy conversion and the transport and break down of carbohydrates and amino acids."
Generating sequence data from the DNA present in a microbial community is useful and worth doing. It provides information about what genes are present and about the metabolic potential of the community. But in isolation, it provides no information about what metabolic activities are actually occurring. The only real way to know what activities are occurring is to measure the activity directly There are practical limitations to doing direct measurements of all enzymatic activities of potential interest so the genetic data is very valuable but its limitations are worth keeping in mind.
Sunday, May 27, 2007
The name
I chose the name Mixotrophy because it suggests the blog will contain a varied diet of thoughts and ideas. Some original and some recycled.
Within biology, the term is used in at least two distinct ways. The more common usage (and the first one I encountered in my studies) describes a large, diverse group of photosynthetic organisms that are capable of acquiring energy from sunlight (autotrophy) and from the degradation of preformed organic compounds (heterotrophy). This definition from the Smithsonian Environmental Research Center gives an idea of how broadly the term is applied.
The other usage is limited to prokaryotes* and is much more specific in its meaning. Here, mixotrophy refers to organisms that are capable of acquiring energy from the oxidation of inorganic compound but are unable to fix carbon. This means they must obtain organic carbon for biosynthesis. A relatively well know example of an organism in this group is Beggiatoa
*Yes I am going to continue using the term and, at the risk of contributing nothing of substance to the discussion, I will probably put up a post about it later in the week
Within biology, the term is used in at least two distinct ways. The more common usage (and the first one I encountered in my studies) describes a large, diverse group of photosynthetic organisms that are capable of acquiring energy from sunlight (autotrophy) and from the degradation of preformed organic compounds (heterotrophy). This definition from the Smithsonian Environmental Research Center gives an idea of how broadly the term is applied.
Mixotrophic organisms gain their nutrition through a combination of photosynthesis and uptake of dissolved or particulate organic material. However, they vary widely in their photosynthetic and heterotrophic capabilities. Some mixotrophs are mainly photosynthetic and only occasionally use an organic energy source. Others meet most of their nutritional demand by phagotrophy, but may use some of the products of photosynthesis from sequestered prey chloroplasts.
The other usage is limited to prokaryotes* and is much more specific in its meaning. Here, mixotrophy refers to organisms that are capable of acquiring energy from the oxidation of inorganic compound but are unable to fix carbon. This means they must obtain organic carbon for biosynthesis. A relatively well know example of an organism in this group is Beggiatoa
*Yes I am going to continue using the term and, at the risk of contributing nothing of substance to the discussion, I will probably put up a post about it later in the week
Saturday, May 26, 2007
Thank you evolution
While a substantial amount of human ingenuity has gone into developing the molecular techniques employed in modern biology, all of these tools have at their root genes and gene products provided to us by the organisms we study. The DNA polymerase used in PCR was isolated from a hyperthermophilic bacteria, the reverse transcriptases used to make expression libraries come courtesy of RNA viruses and the multitude of cloning vectors, transposons and antibiotic cassettes are all derived from DNA isolated from natural sources. Other examples are luminescent compounds from fireflies or soft corals and fluorescent proteins from jellyfish.
Friday, May 25, 2007
ASM general meeting
I have been in Toronto all week at the annual American Society for Microbiology meeting. These big meetings can be a bit overwhelming given the numerous concurrent talks and poster sessions running every day. In reflecting on the many topics I learned about (some new and some quite familiar to me), I am struck by how ubiquitous the use of molecular techniques has become. When the information generated with these techniques is combined with biochemical and physiological data, incredible insight can be gained into the mechanisms by which microbes survive and thrive in diverse environments.
Examples include relatively old techniques such as the generation of mutant strains by random or site directed mutagenesis and the rescuing of these mutants by complementation. This reductionist approach provides information about the role specific genes play in the survival of microbes in different environments, in the utilization of specific food sources or the resistance to stresses.
Newer innovations such as 454 sequencing combined with modern computing power and bioinformatics software allow researchers to approach the same questions more broadly. For example, the technique can be used to sequence the entire genome of multiple strains of the same species of bacteria or to generate enormous databases of the specific genes expressed by microbes grown under specific culture conditions.
These approaches are revolutionizing microbial ecology.
Examples include relatively old techniques such as the generation of mutant strains by random or site directed mutagenesis and the rescuing of these mutants by complementation. This reductionist approach provides information about the role specific genes play in the survival of microbes in different environments, in the utilization of specific food sources or the resistance to stresses.
Newer innovations such as 454 sequencing combined with modern computing power and bioinformatics software allow researchers to approach the same questions more broadly. For example, the technique can be used to sequence the entire genome of multiple strains of the same species of bacteria or to generate enormous databases of the specific genes expressed by microbes grown under specific culture conditions.
These approaches are revolutionizing microbial ecology.
Thursday, May 24, 2007
Welcome
Welcome to mixotrophy. I set of the template quite a while ago but with all the interesting blogs out there that I spend time reading and commenting on, I have not found the time to post anything here. Hopefully that will change. Please check back soon
Andrew
Andrew
Subscribe to:
Posts (Atom)



