Chloroplasts, like mitochondria, have their own chromosomal DNA. This is, of course, evidence for the endosymbiotic theory of the origin of chloroplasts. It is also useful because it allows researchers to use the DNA to identify the source of the chloroplasts present in kleptoplastic organisms. This is a fairly standard method and is the way that Gast et al 2007 determined the source of the chloroplasts in the Antartic dinoflagellates discussed in a previous post.
Chloroplasts, like mitochondria, have their own chromosomal DNA. This is, of course, evidence for the endosymbiotic theory of the origin of chloroplasts. It is also useful because it allows researchers to use the DNA to identify the source of the chloroplasts present in kleptoplastic organisms. This is a fairly standard method and is the way that Gast et al 2007 determined the source of the chloroplasts in the Antartic dinoflagellates discussed in a previous post.In a paper (pdf) a few years ago in Limnology and Oceanography, McManus et al. used the chloroplasts present in the kleptoplastic tide-pool ciliates, Strombidium oculatum and Strombidium styliferseen to help reveal an interesting life history. The chloroplasts were from the large multicellular macroalgae Enteromorpha clathrata which raised the question of how these unicellular cilliates were able to acquire macroalgal chloroplasts.
McManus et al. found that the cilliates don't appear to be grazing directly on the large strands of the mature algae but on the small motile reproductive cells, called zoospores, mature algal strands release.
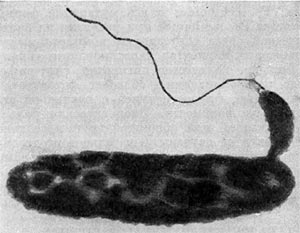
In addition to chloroplasts, the zoospores contain a pigmented eyespot. As the photo above (from the paper's figure 1) demonstrates, the kleptoplastic cilliates contain a pigmented eyespot similar to the ones possessed by the zoospores. This suggests that the Strombidium cilliates also owe their phototaxic abilities to the alga cells they ingest.
Some other interesting points about these cilliates:
- They are tidal organisms and live by the rhythm of the tides, becoming active during low tide when tidal pools are calm, and then attach to surfaces and encyst during high tide, presumably to prevent them from being washed out to sea and away from their food.
- They appear to be obligate mixotrophs, unable to grow in the dark or in the absence of algal food.
McManus, G. B., H. Zhang, and S. Lin. 2004. Marine planktonic ciliates that prey on macroalgae and enslave their chloroplasts. Limnol. Oceanogr. 49:308-313.